Code
library(plotly)
library(gganimate)
library(knitr)
library(kableExtra)
library(tidybayes)
library(posterior)
library(bayesplot)
library(cmdstanr)
library(tidyverse)Stay tuned for (ir)regular updates!
Stan is not specialized for pharmacometrics, so it does not automatically handle dosing events and common models that pharmacometrics-specific software like NONMEM, Monolix, Pumas, and nlmixr do. These limitations are mitigated when Stan is used in conjunction with Torsten, a collection of Stan functions that are analogous to NONMEM’s ADVANs. Given a dataset that follows NONMEM’s NM-TRAN specifications, these functions specify common PMx models and do all the event handling required to fit (or simulate) pharmacometric data.
Since we have the ability to fit standard NONMEM datasets, we can just read in a typical NONMEM-ready dataset that we get from our programming groups. It will then require a small amount of wrangling to get ready for Stan.
nonmem_data <- read_csv(here::here("Data/depot_1cmt_ppa_covariates.csv"),
na = ".") %>%
rename_all(tolower) %>%
rename(ID = "id",
DV = "dv") %>%
mutate(DV = if_else(is.na(DV), 5555555, DV), # This value can be anything except NA. It'll be indexed away
bloq = if_else(is.na(bloq), -999, bloq)) # This value can be anything except NA. It'll be indexed away
nonmem_data %>%
mutate(across(where(is.numeric), \(x) round(x, 3))) %>%
DT::datatable()(p_1 <- ggplot(nonmem_data %>%
group_by(ID) %>%
mutate(Dose = factor(max(amt, na.rm = TRUE))) %>%
ungroup() %>%
filter(mdv == 0)) +
geom_line(mapping = aes(x = time, y = DV, group = ID, color = Dose)) +
geom_point(mapping = aes(x = time, y = DV, group = ID, color = Dose)) +
scale_color_discrete(name = "Dose (mg)") +
scale_y_continuous(name = latex2exp::TeX("$Drug\\;Conc.\\;(\\mu g/mL)$"),
limits = c(NA, NA),
trans = "log10") +
scale_x_continuous(name = "Time (d)",
breaks = seq(0, 216, by = 24),
labels = seq(0, 216/24, by = 24/24),
limits = c(0, NA)) +
theme_bw(18) +
theme(axis.text = element_text(size = 14, face = "bold"),
axis.title = element_text(size = 18, face = "bold"),
axis.line = element_line(linewidth = 2),
legend.position = "bottom"))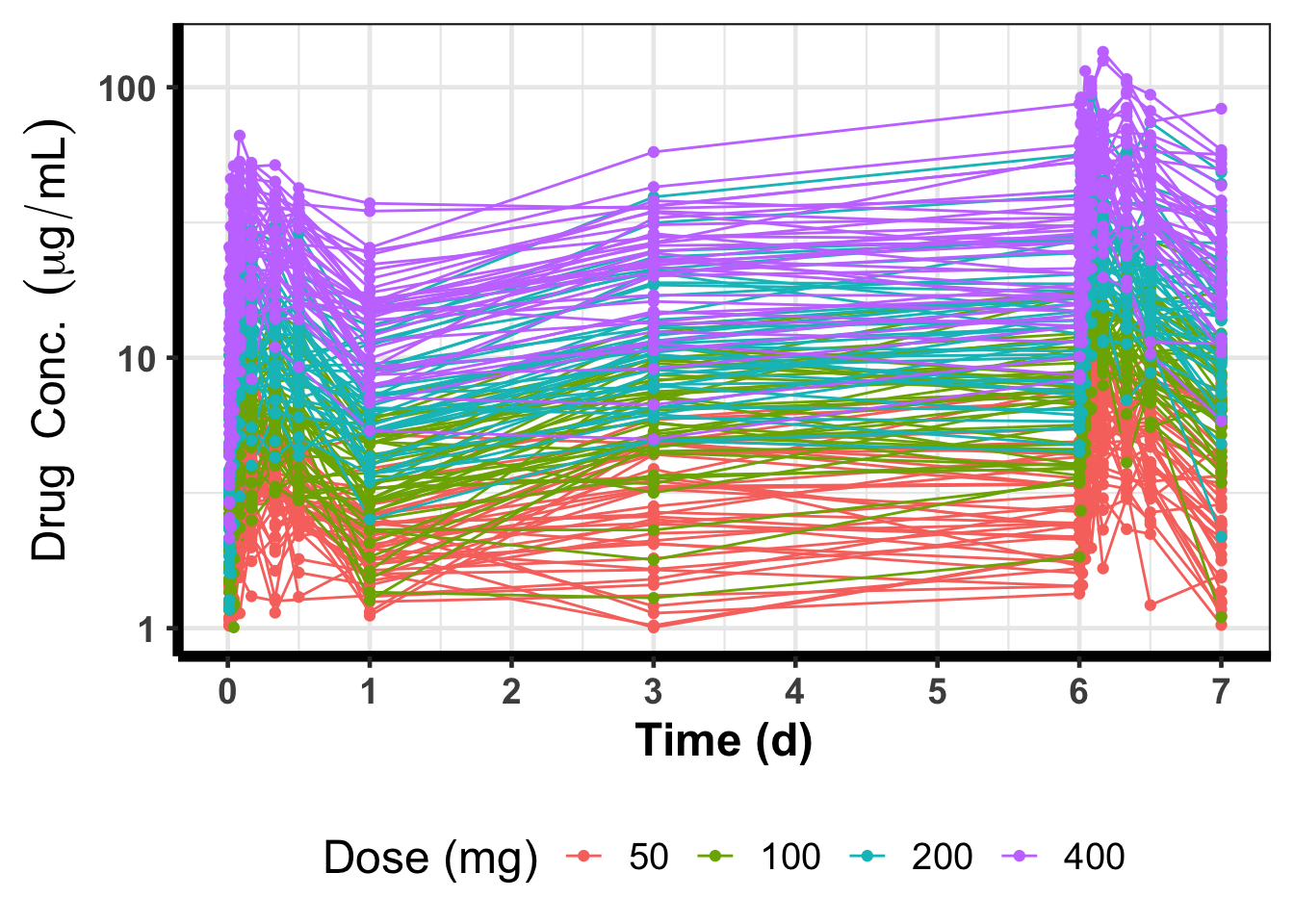
Then we need to wrangle the data to get it ready to be input into Stan:
n_subjects <- nonmem_data %>% # number of individuals
distinct(ID) %>%
count() %>%
deframe()
n_total <- nrow(nonmem_data) # total number of records
i_obs <- nonmem_data %>% # indices of the observations
mutate(row_num = 1:n()) %>%
filter(evid == 0) %>%
select(row_num) %>%
deframe()
n_obs <- length(i_obs) # number of observations
subj_start <- nonmem_data %>% # Indices that indicate which row is the start of a new subject
mutate(row_num = 1:n()) %>%
group_by(ID) %>%
slice_head(n = 1) %>%
ungroup() %>%
select(row_num) %>%
deframe()
subj_end <- c(subj_start[-1] - 1, n_total) # Indices that indicate which row is the end of a subject
# Covariates
wt <- nonmem_data %>%
group_by(ID) %>%
distinct(wt) %>%
ungroup() %>%
pull(wt)
cmppi <- nonmem_data %>%
group_by(ID) %>%
distinct(cmppi) %>%
ungroup() %>%
pull(cmppi)
egfr <- nonmem_data %>%
group_by(ID) %>%
distinct(egfr) %>%
ungroup() %>%
pull(egfr)
race <- nonmem_data %>%
group_by(ID) %>%
distinct(race) %>%
ungroup() %>%
pull(race)
n_races <- length(unique(race))Here’s the Stan model:
#| code-fold: false
// First Order Absorption (oral/subcutaneous)
// One-compartment PK Model
// IIV on CL, VC, and Ka (full covariance matrix)
// proportional plus additive error - DV = IPRED*(1 + eps_p) + eps_a
// Matrix-exponential solution using Torsten (the matrix-exponential seems to be
// faster than the analytical solution for this model)
// Implements threading for within-chain parallelization
// Deals with BLOQ values by the "CDF trick" (M4)
// Since we have a normal distribution on the error, but the DV must be > 0, it
// truncates the likelihood below at 0
// For PPC, it generates values from a normal that is truncated below at 0
// Covariates:
// 1) Body Weight on CL and VC - (wt/70)^theta
// 2) Concomitant administration of protein pump inhibitors (CMPPI)
// on KA (0/1) - exp(theta*cmppi)
// 3) eGFR on CL (continuous) - (eGFR/90)^theta
// 4) Race on VC - exp(theta_vc_{race}*I(race == {race}))
functions{
int num_between(int lb, int ub, array[] int y){
int n = 0;
for(i in 1:num_elements(y)){
if(y[i] >= lb && y[i] <= ub)
n = n + 1;
}
return n;
}
array[] int find_between(int lb, int ub, array[] int y) {
// vector[num_between(lb, ub, y)] result;
array[num_between(lb, ub, y)] int result;
int n = 1;
for (i in 1:num_elements(y)) {
if (y[i] >= lb && y[i] <= ub) {
result[n] = y[i];
n = n + 1;
}
}
return result;
}
vector find_between_vec(int lb, int ub, array[] int idx, vector y) {
vector[num_between(lb, ub, idx)] result;
int n = 1;
if(num_elements(idx) != num_elements(y)) reject("illegal input");
for (i in 1:rows(y)) {
if (idx[i] >= lb && idx[i] <= ub) {
result[n] = y[i];
n = n + 1;
}
}
return result;
}
real normal_lb_rng(real mu, real sigma, real lb){
real p_lb = normal_cdf(lb | mu, sigma);
real u = uniform_rng(p_lb, 1);
real y = mu + sigma * inv_Phi(u);
return y;
}
real partial_sum_lpmf(array[] int seq_subj, int start, int end,
vector dv_obs, array[] int dv_obs_id, array[] int i_obs,
array[] real amt, array[] int cmt, array[] int evid,
array[] real time, array[] real rate, array[] real ii,
array[] int addl, array[] int ss,
array[] int subj_start, array[] int subj_end,
vector CL, vector VC, vector KA,
real sigma_sq_p, real sigma_sq_a, real sigma_p_a,
vector lloq, array[] int bloq,
int n_random, int n_subjects, int n_total,
array[] real bioav, array[] real tlag, int n_cmt){
real ptarget = 0;
int N = end - start + 1; // number of subjects in this slice
vector[n_total] dv_ipred;
matrix[n_total, 2] x_ipred;
int n_obs_slice = num_between(subj_start[start], subj_end[end], i_obs);
array[n_obs_slice] int i_obs_slice = find_between(subj_start[start],
subj_end[end], i_obs);
vector[n_obs_slice] dv_obs_slice = find_between_vec(start, end,
dv_obs_id, dv_obs);
vector[n_obs_slice] ipred_slice;
vector[n_obs_slice] lloq_slice = lloq[i_obs_slice];
array[n_obs_slice] int bloq_slice = bloq[i_obs_slice];
for(n in 1:N){ // loop over subjects in this slice
int j = n + start - 1; // j is the ID of the current subject
matrix[n_cmt, n_cmt] K = rep_matrix(0, n_cmt, n_cmt);
K[1, 1] = -KA[j];
K[2, 1] = KA[j];
K[2, 2] = -CL[j]/VC[j];
x_ipred[subj_start[j]:subj_end[j], ] =
pmx_solve_linode(time[subj_start[j]:subj_end[j]],
amt[subj_start[j]:subj_end[j]],
rate[subj_start[j]:subj_end[j]],
ii[subj_start[j]:subj_end[j]],
evid[subj_start[j]:subj_end[j]],
cmt[subj_start[j]:subj_end[j]],
addl[subj_start[j]:subj_end[j]],
ss[subj_start[j]:subj_end[j]],
K, bioav, tlag)';
dv_ipred[subj_start[j]:subj_end[j]] =
x_ipred[subj_start[j]:subj_end[j], 2] ./ VC[j];
}
ipred_slice = dv_ipred[i_obs_slice];
for(i in 1:n_obs_slice){
real ipred_tmp = ipred_slice[i];
real sigma_tmp = sqrt(square(ipred_tmp) * sigma_sq_p + sigma_sq_a +
2*ipred_tmp*sigma_p_a);
if(bloq_slice[i] == 1){
ptarget += log_diff_exp(normal_lcdf(lloq_slice[i] | ipred_tmp,
sigma_tmp),
normal_lcdf(0.0 | ipred_tmp, sigma_tmp)) -
normal_lccdf(0.0 | ipred_tmp, sigma_tmp);
}else{
ptarget += normal_lpdf(dv_obs_slice[i] | ipred_tmp, sigma_tmp) -
normal_lccdf(0.0 | ipred_tmp, sigma_tmp);
}
}
return ptarget;
}
}
data{
int n_subjects;
int n_total;
int n_obs;
array[n_obs] int i_obs;
array[n_total] int ID;
array[n_total] real amt;
array[n_total] int cmt;
array[n_total] int evid;
array[n_total] real rate;
array[n_total] real ii;
array[n_total] int addl;
array[n_total] int ss;
array[n_total] real time;
vector<lower = 0>[n_total] dv;
array[n_subjects] int subj_start;
array[n_subjects] int subj_end;
vector[n_total] lloq;
array[n_total] int bloq;
array[n_subjects] real<lower = 0> wt; // baseline bodyweight (kg)
array[n_subjects] int<lower = 0, upper = 1> cmppi; // cmppi
array[n_subjects] real<lower = 0> egfr; // eGFR
int<lower = 2> n_races; // number of unique races
array[n_subjects] int<lower = 1, upper = n_races> race; // race
real<lower = 0> location_tvcl; // Prior Location parameter for CL
real<lower = 0> location_tvvc; // Prior Location parameter for VC
real<lower = 0> location_tvka; // Prior Location parameter for KA
real<lower = 0> scale_tvcl; // Prior Scale parameter for CL
real<lower = 0> scale_tvvc; // Prior Scale parameter for VC
real<lower = 0> scale_tvka; // Prior Scale parameter for KA
real<lower = 0> scale_omega_cl; // Prior scale parameter for omega_cl
real<lower = 0> scale_omega_vc; // Prior scale parameter for omega_vc
real<lower = 0> scale_omega_ka; // Prior scale parameter for omega_ka
real<lower = 0> lkj_df_omega; // Prior degrees of freedom for omega cor mat
real<lower = 0> scale_sigma_p; // Prior Scale parameter for proportional error
real<lower = 0> scale_sigma_a; // Prior Scale parameter for additive error
real<lower = 0> lkj_df_sigma; // Prior degrees of freedom for sigma cor mat
int<lower = 0, upper = 1> prior_only; // Want to simulate from the prior?
int<lower = 0, upper = prior_only> no_gq_predictions; // Leave out PREDS and IPREDS in
// generated quantities. Useful
// for simulating prior parameters
// but don't want prior predictions
}
transformed data{
int grainsize = 1;
vector<lower = 0>[n_obs] dv_obs = dv[i_obs];
array[n_obs] int dv_obs_id = ID[i_obs];
vector[n_obs] lloq_obs = lloq[i_obs];
array[n_obs] int bloq_obs = bloq[i_obs];
int n_random = 3; // Number of random effects
int n_cmt = 2; // Number of states in the ODEs
array[n_random] real scale_omega = {scale_omega_cl, scale_omega_vc,
scale_omega_ka};
array[2] real scale_sigma = {scale_sigma_p, scale_sigma_a};
array[n_subjects] int seq_subj = linspaced_int_array(n_subjects, 1, n_subjects); // reduce_sum over subjects
array[n_cmt] real bioav = rep_array(1.0, n_cmt); // Hardcoding, but could be data or a parameter in another situation
array[n_cmt] real tlag = rep_array(0.0, n_cmt);
}
parameters{
real<lower = 0> TVCL;
real<lower = 0> TVVC;
real<lower = TVCL/TVVC> TVKA;
real theta_cl_wt;
real theta_vc_wt;
real theta_ka_cmppi;
real theta_cl_egfr;
real theta_vc_race2;
real theta_vc_race3;
real theta_vc_race4;
vector<lower = 0>[n_random] omega;
cholesky_factor_corr[n_random] L;
vector<lower = 0>[2] sigma;
cholesky_factor_corr[2] L_Sigma;
matrix[n_random, n_subjects] Z;
}
transformed parameters{
vector[n_subjects] eta_cl;
vector[n_subjects] eta_vc;
vector[n_subjects] eta_ka;
vector[n_subjects] CL;
vector[n_subjects] VC;
vector[n_subjects] KA;
vector[n_subjects] KE;
real<lower = 0> sigma_p = sigma[1];
real<lower = 0> sigma_a = sigma[2];
real<lower = 0> sigma_sq_p = square(sigma_p);
real<lower = 0> sigma_sq_a = square(sigma_a);
real cor_p_a;
real sigma_p_a;
{
row_vector[n_random] typical_values = to_row_vector({TVCL, TVVC, TVKA});
matrix[n_subjects, n_random] eta = diag_pre_multiply(omega, L * Z)';
matrix[n_subjects, n_random] theta =
(rep_matrix(typical_values, n_subjects) .* exp(eta));
vector[n_races] theta_vc_race = [0, theta_vc_race2, theta_vc_race3,
theta_vc_race4]';
matrix[2, 2] R_Sigma = multiply_lower_tri_self_transpose(L_Sigma);
matrix[2, 2] Sigma = quad_form_diag(R_Sigma, sigma);
eta_cl = col(eta, 1);
eta_vc = col(eta, 2);
eta_ka = col(eta, 3);
cor_p_a = R_Sigma[1, 2];
sigma_p_a = Sigma[1, 2];
for(j in 1:n_subjects){
real wt_over_70 = wt[j]/70;
real wt_adjustment_cl = wt_over_70^theta_cl_wt;
real wt_adjustment_vc = wt_over_70^theta_vc_wt;
real cmppi_adjustment_ka = exp(theta_ka_cmppi*cmppi[j]);
real egfr_adjustment_cl = (egfr[j]/90)^theta_cl_egfr;
real race_adjustment_vc = exp(theta_vc_race[race[j]]);
row_vector[n_random] theta_j = theta[j]; // access the parameters for subject j
CL[j] = theta_j[1] * wt_adjustment_cl * egfr_adjustment_cl;
VC[j] = theta_j[2] * wt_adjustment_vc * race_adjustment_vc;
KA[j] = theta_j[3] * cmppi_adjustment_ka;
KE[j] = CL[j]/VC[j];
}
}
}
model{
// Priors
TVCL ~ lognormal(log(location_tvcl), scale_tvcl);
TVVC ~ lognormal(log(location_tvvc), scale_tvvc);
TVKA ~ lognormal(log(location_tvka), scale_tvka) T[TVCL/TVVC, ];
theta_cl_wt ~ std_normal();
theta_vc_wt ~ std_normal();
theta_ka_cmppi ~ std_normal();
theta_cl_egfr ~ std_normal();
theta_vc_race2 ~ std_normal();
theta_vc_race3 ~ std_normal();
theta_vc_race4 ~ std_normal();
omega ~ normal(0, scale_omega);
L ~ lkj_corr_cholesky(lkj_df_omega);
sigma ~ normal(0, scale_sigma);
L_Sigma ~ lkj_corr_cholesky(lkj_df_sigma);
to_vector(Z) ~ std_normal();
// Likelihood
if(prior_only == 0){
target += reduce_sum(partial_sum_lupmf, seq_subj, grainsize,
dv_obs, dv_obs_id, i_obs,
amt, cmt, evid, time,
rate, ii, addl, ss, subj_start, subj_end,
CL, VC, KA,
sigma_sq_p, sigma_sq_a, sigma_p_a,
lloq, bloq,
n_random, n_subjects, n_total,
bioav, tlag, n_cmt);
}
}
generated quantities{
real<lower = 0> omega_cl = omega[1];
real<lower = 0> omega_vc = omega[2];
real<lower = 0> omega_ka = omega[3];
real<lower = 0> omega_sq_cl = square(omega_cl);
real<lower = 0> omega_sq_vc = square(omega_vc);
real<lower = 0> omega_sq_ka = square(omega_ka);
real cor_cl_vc;
real cor_cl_ka;
real cor_vc_ka;
real omega_cl_vc;
real omega_cl_ka;
real omega_vc_ka;
vector[no_gq_predictions ? 0 : n_obs] pred;
vector[no_gq_predictions ? 0 : n_obs] epred_stan;
vector[no_gq_predictions ? 0 : n_obs] ipred;
vector[no_gq_predictions ? 0 : n_obs] epred;
vector[no_gq_predictions ? 0 : n_obs] dv_ppc;
vector[no_gq_predictions ? 0 : n_obs] log_lik;
vector[no_gq_predictions ? 0 : n_obs] iwres;
{
matrix[n_random, n_random] R = multiply_lower_tri_self_transpose(L);
matrix[n_random, n_random] Omega = quad_form_diag(R, omega);
cor_cl_vc = R[1, 2];
cor_cl_ka = R[1, 3];
cor_vc_ka = R[2, 3];
omega_cl_vc = Omega[1, 2];
omega_cl_ka = Omega[1, 3];
omega_vc_ka = Omega[2, 3];
}
if(no_gq_predictions == 0){
vector[n_subjects] CL_new;
vector[n_subjects] VC_new;
vector[n_subjects] KA_new;
vector[n_subjects] KE_new;
vector[n_total] dv_pred;
matrix[n_total, n_cmt] x_pred;
vector[n_total] dv_epred;
matrix[n_total, n_cmt] x_epred;
vector[n_total] dv_ipred;
matrix[n_total, n_cmt] x_ipred;
matrix[n_subjects, n_random] eta_new;
matrix[n_subjects, n_random] theta_new;
vector[n_races] theta_vc_race = [0, theta_vc_race2, theta_vc_race3,
theta_vc_race4]';
for(i in 1:n_subjects){
eta_new[i, ] = multi_normal_cholesky_rng(rep_vector(0, n_random),
diag_pre_multiply(omega, L))';
}
theta_new = (rep_matrix(to_row_vector({TVCL, TVVC, TVKA}), n_subjects) .* exp(eta_new));
for(j in 1:n_subjects){
real wt_over_70 = wt[j]/70;
real wt_adjustment_cl = wt_over_70^theta_cl_wt;
real wt_adjustment_vc = wt_over_70^theta_vc_wt;
real cmppi_adjustment_ka = exp(theta_ka_cmppi*cmppi[j]);
real egfr_adjustment_cl = (egfr[j]/90)^theta_cl_egfr;
real race_adjustment_vc = exp(theta_vc_race[race[j]]);
row_vector[n_random] theta_j_new = theta_new[j]; // access the parameters for subject j
CL_new[j] = theta_j_new[1] * wt_adjustment_cl * egfr_adjustment_cl;
VC_new[j] = theta_j_new[2] * wt_adjustment_vc * race_adjustment_vc;
KA_new[j] = theta_j_new[3] * cmppi_adjustment_ka;
KE_new[j] = CL_new[j]/VC_new[j];
real cl_p = TVCL * wt_adjustment_cl * egfr_adjustment_cl;
real vc_p = TVVC * wt_adjustment_vc * race_adjustment_vc;
real ka_p = TVKA * cmppi_adjustment_ka;
matrix[n_cmt, n_cmt] K = rep_matrix(0, n_cmt, n_cmt);
matrix[n_cmt, n_cmt] K_epred = rep_matrix(0, n_cmt, n_cmt);
matrix[n_cmt, n_cmt] K_p = rep_matrix(0, n_cmt, n_cmt);
K[1, 1] = -KA[j];
K[2, 1] = KA[j];
K[2, 2] = -KE[j];
x_ipred[subj_start[j]:subj_end[j], ] =
pmx_solve_linode(time[subj_start[j]:subj_end[j]],
amt[subj_start[j]:subj_end[j]],
rate[subj_start[j]:subj_end[j]],
ii[subj_start[j]:subj_end[j]],
evid[subj_start[j]:subj_end[j]],
cmt[subj_start[j]:subj_end[j]],
addl[subj_start[j]:subj_end[j]],
ss[subj_start[j]:subj_end[j]],
K, bioav, tlag)';
K_epred[1, 1] = -KA_new[j];
K_epred[2, 1] = KA_new[j];
K_epred[2, 2] = -KE_new[j];
x_epred[subj_start[j]:subj_end[j], ] =
pmx_solve_linode(time[subj_start[j]:subj_end[j]],
amt[subj_start[j]:subj_end[j]],
rate[subj_start[j]:subj_end[j]],
ii[subj_start[j]:subj_end[j]],
evid[subj_start[j]:subj_end[j]],
cmt[subj_start[j]:subj_end[j]],
addl[subj_start[j]:subj_end[j]],
ss[subj_start[j]:subj_end[j]],
K_epred, bioav, tlag)';
K_p[1, 1] = -ka_p;
K_p[2, 1] = ka_p;
K_p[2, 2] = -cl_p/vc_p;
x_pred[subj_start[j]:subj_end[j],] =
pmx_solve_linode(time[subj_start[j]:subj_end[j]],
amt[subj_start[j]:subj_end[j]],
rate[subj_start[j]:subj_end[j]],
ii[subj_start[j]:subj_end[j]],
evid[subj_start[j]:subj_end[j]],
cmt[subj_start[j]:subj_end[j]],
addl[subj_start[j]:subj_end[j]],
ss[subj_start[j]:subj_end[j]],
K_p, bioav, tlag)';
dv_ipred[subj_start[j]:subj_end[j]] =
x_ipred[subj_start[j]:subj_end[j], 2] ./ VC[j];
dv_epred[subj_start[j]:subj_end[j]] =
x_epred[subj_start[j]:subj_end[j], 2] ./ VC_new[j];
dv_pred[subj_start[j]:subj_end[j]] =
x_pred[subj_start[j]:subj_end[j], 2] ./ vc_p;
}
pred = dv_pred[i_obs];
epred_stan = dv_epred[i_obs];
ipred = dv_ipred[i_obs];
for(i in 1:n_obs){
real ipred_tmp = ipred[i];
real sigma_tmp = sqrt(square(ipred_tmp) * sigma_sq_p + sigma_sq_a +
2*ipred_tmp*sigma_p_a);
real epred_tmp = epred_stan[i];
real sigma_tmp_e = sqrt(square(epred_tmp) * sigma_sq_p + sigma_sq_a +
2*epred_tmp*sigma_p_a);
dv_ppc[i] = normal_lb_rng(ipred_tmp, sigma_tmp, 0.0);
epred[i] = normal_lb_rng(epred_tmp, sigma_tmp_e, 0.0);
if(bloq_obs[i] == 1){
// log_lik[i] = log(normal_cdf(lloq_obs[i] | ipred_tmp, sigma_tmp) -
// normal_cdf(0.0 | ipred_tmp, sigma_tmp)) -
// normal_lccdf(0.0 | ipred_tmp, sigma_tmp);
log_lik[i] = log_diff_exp(normal_lcdf(lloq_obs[i] | ipred_tmp, sigma_tmp),
normal_lcdf(0.0 | ipred_tmp, sigma_tmp)) -
normal_lccdf(0.0 | ipred_tmp, sigma_tmp);
}else{
log_lik[i] = normal_lpdf(dv_obs[i] | ipred_tmp, sigma_tmp) -
normal_lccdf(0.0 | ipred_tmp, sigma_tmp);
}
iwres[i] = (dv_obs[i] - ipred_tmp)/sigma_tmp;
}
}
}
model <- cmdstan_model(here::here("Stan/Fit/depot_1cmt_ppa_covariates.stan"),
cpp_options = list(stan_threads = TRUE))
stan_data <- list(n_subjects = n_subjects,
n_total = n_total,
n_obs = n_obs,
i_obs = i_obs,
ID = nonmem_data$ID,
amt = nonmem_data$amt,
cmt = nonmem_data$cmt,
evid = nonmem_data$evid,
rate = nonmem_data$rate,
ii = nonmem_data$ii,
addl = nonmem_data$addl,
ss = nonmem_data$ss,
time = nonmem_data$time,
dv = nonmem_data$DV,
subj_start = subj_start,
subj_end = subj_end,
wt = wt,
cmppi = cmppi,
egfr = egfr,
n_races = n_races,
race = race,
lloq = nonmem_data$lloq,
bloq = nonmem_data$bloq,
location_tvcl = 0.5,
location_tvvc = 4,
location_tvka = 0.8,
scale_tvcl = 1,
scale_tvvc = 1,
scale_tvka = 1,
scale_omega_cl = 0.4,
scale_omega_vc = 0.4,
scale_omega_ka = 0.4,
lkj_df_omega = 2,
scale_sigma_p = 0.5,
scale_sigma_a = 0.5,
lkj_df_sigma = 2,
prior_only = 0,
no_gq_predictions = 0)
fit <- model$sample(data = stan_data,
seed = 112358,
chains = 4,
parallel_chains = 4,
threads_per_chain = parallel::detectCores()/4,
iter_warmup = 500,
iter_sampling = 1000,
adapt_delta = 0.8,
refresh = 500,
max_treedepth = 10,
# output_dir = "depot_1cmt_linear_covariates/Stan/Fits/Output",
# output_basename = "ppa_covariates",
init = function() list(TVCL = rlnorm(1, log(1), 0.3),
TVVC = rlnorm(1, log(8), 0.3),
TVKA = rlnorm(1, log(0.8), 0.3),
theta_cl_wt = rnorm(1, 0, 0.2),
theta_vc_wt = rnorm(1, 0, 0.2),
theta_ka_cmppi = rnorm(1, 0, 0.2),
theta_cl_egfr = rnorm(1, 0, 0.2),
theta_vc_race2 = rnorm(1, 0, 0.2),
theta_vc_race3 = rnorm(1, 0, 0.2),
theta_vc_race4 = rnorm(1, 0, 0.2),
omega = rlnorm(3, log(0.3), 0.3),
sigma = rlnorm(2, log(0.2), 0.3)))
fit$save_object(
"Stan/Fits/depot_1cmt_ppa_covariates.rds")
fit$save_data_file(dir = "Stan/Fits/Stan_Data",
basename = "ppa_covariates", timestamp = FALSE,
random = FALSE)